Why Use OOP?
R is traditionally a functional programming language, meaning that when we code in R, we tend to think in terms of functions. For instance, given a data set, what function can I create to plot a certain variable? Object oriented programming (OOP) introduces the concept of “classes.” While classes are used extensively in other programming languages such as Java, their use in R differs from conventional uses. In part, this is because R has several unique OOP systems which differ from one another in key ways. The three most popular systems are S3, S4, and R6. While different R communities leverage different systems, we will focus on S3, which is favored by the folks over at RStudio, the Tidyverse, and other related organizations. For more information, see the additional reading section below.
An Example Use Case
Consider the case where you want to create a function that works for a specific type of data format. Perhaps there you are interested in a specific column and want to ensure your data has this column present. For instance, let us take a look at the following dataset.
#> # A tibble: 832 × 10
#> sample gene_id gene mutation_name exonic_func aa_change targeted
#> <chr> <chr> <chr> <chr> <chr> <chr> <chr>
#> 1 D10-JJJ-23 PF3D7_0106300 atp6 atp6-Ala623Glu missense_va… Ala623Glu Yes
#> 2 D10-JJJ-43 PF3D7_0106300 atp6 atp6-Ala623Glu missense_va… Ala623Glu Yes
#> 3 D10-JJJ-55 PF3D7_0106300 atp6 atp6-Ala623Glu missense_va… Ala623Glu Yes
#> 4 D10-JJJ-5 PF3D7_0106300 atp6 atp6-Ala623Glu missense_va… Ala623Glu Yes
#> 5 D10-JJJ-47 PF3D7_0106300 atp6 atp6-Ala623Glu missense_va… Ala623Glu Yes
#> 6 D10-JJJ-15 PF3D7_0106300 atp6 atp6-Ala623Glu missense_va… Ala623Glu Yes
#> 7 D10-JJJ-27 PF3D7_0106300 atp6 atp6-Ala623Glu missense_va… Ala623Glu Yes
#> 8 D10-JJJ-10 PF3D7_0106300 atp6 atp6-Ala623Glu missense_va… Ala623Glu Yes
#> 9 D10-JJJ-28 PF3D7_0106300 atp6 atp6-Ala623Glu missense_va… Ala623Glu Yes
#> 10 D10-JJJ-52 PF3D7_0106300 atp6 atp6-Ala623Glu missense_va… Ala623Glu Yes
#> # … with 822 more rows, and 3 more variables: ref_umi_count <dbl>,
#> # alt_umi_count <dbl>, coverage <dbl>
Let us say we want to create a function mutation_prevalence which determines
the prevalence of each mutation in the dataset. Moreover, we would like to plot
this prevalence.
prevalence <- mutation_prevalence(data, 5)
prevalence
#> # A tibble: 16 × 4
#> mutation_name n_total n_mutant prevalence
#> <chr> <int> <int> <dbl>
#> 1 atp6-Ala623Glu 36 NA NA
#> 2 atp6-Glu431Lys 39 NA NA
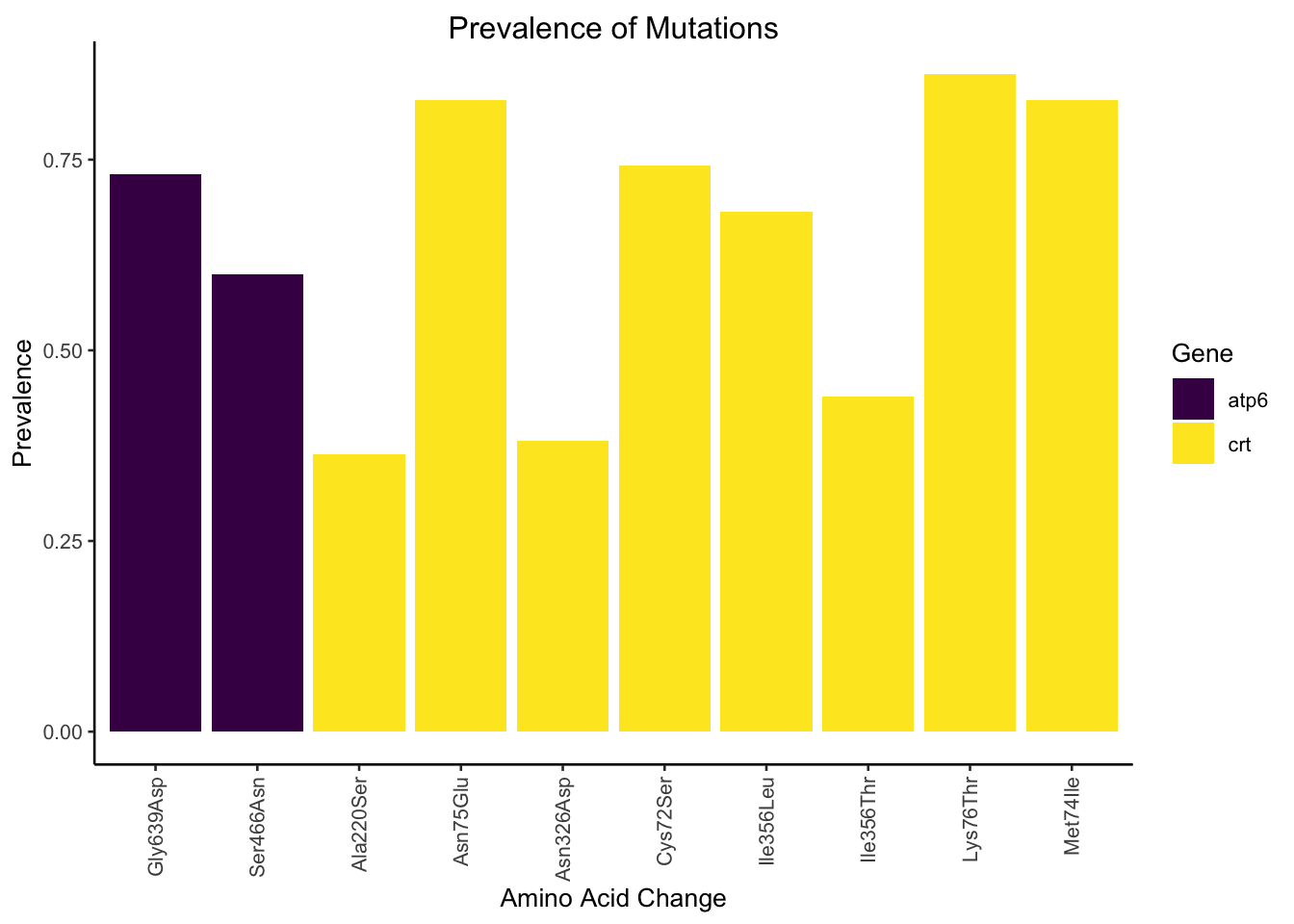
#> 3 atp6-Gly639Asp 26 19 0.731
#> 4 atp6-Ser466Asn 15 9 0.6
#> 5 atp6-Ser769Asn 17 NA NA
#> 6 crt-Ala220Ser 11 4 0.364
#> 7 crt-Asn326Asp 21 8 0.381
#> 8 crt-Asn326Ser 26 NA NA
#> 9 crt-Asn75Glu 29 24 0.828
#> 10 crt-Cys72Ser 31 23 0.742
#> 11 crt-His97Leu 47 NA NA
#> 12 crt-His97Tyr 47 NA NA
#> 13 crt-Ile356Leu 22 15 0.682
#> 14 crt-Ile356Thr 41 18 0.439
#> 15 crt-Lys76Thr 29 25 0.862
#> 16 crt-Met74Ile 29 24 0.828
While creating this function may require some thought, it makes intuitive sense
that the result will contain four columns as seen above. Two of these columns,
n_total and n_mutant, are used to compute the final column, prevalence.
Therefore, if we wanted to visualize our data, it is not very important for us
to consider the two columns n_total and n_mutant. Rather, we care about
the mutation_name and the prevalence.
If we were to create an function, plot_prevalence, the easiest way to code it
would be to give the function a data argument:
plot_prevalence <- function(data) {
# code for plotting goes here
}
Notice that in the given format, we could feed any dataset into our function.
In other words, there are no checks to make sure we are feeding in the output
of mutation_prevalence(). How could we address this potential problem?
Potential Solutions
The most obvious solution would be to check that some key columns exist:
plot_prevalence <- function(data) {
if (!"mutation_name" %in% colnames(data)) {
stop("Missing key column!", call. = FALSE)
}
# code for plotting goes here
}
Howerver, another strategy would be to use classes! If we assigned a class, say
mut_prev, to the output of mutation_prevalence(), we could easily check if
the input is of type mut_prev:
plot_prevalence <- function(data) {
if (!inherits(data, "mut_prev")) {
stop("Wrong class input!", call. = FALSE)
}
# code for plotting goes here
}
But Wait, There’s More!
Now, using classes to solve this issue might seem like overkill.
After all, why create an entire new class when you can just have an if()
statement? The real power of object-oriented programming is the ability to use
polymorphism1. What we mean by this is the
ability to use the same function for many types of input. An example of this
behavior is the base R function print() which behaves differently depending on
the class of the input.
# A vector
print(c(1, 2))
#> [1] 1 2
# A tibble
print(tibble::as_tibble(c(1, 2)))
#> # A tibble: 2 × 1
#> value
#> <dbl>
#> 1 1
#> 2 2
Using OOP, developers can customize how certain functions interact with certain
objects. Revisiting our mut_prev class, we could even change the way the table
is printed! Another common example is plotting a dataset. Say we have developed
a package that introduces three different classes of datasets. We can then
create a plot method for each type of class.
plot.class1 <- function(x) {
# plotting code
}
plot.class2 <- function(x) {
# plotting code
}
plot.class3 <- function(x) {
# plotting code
}
We can witness this in action with our class mut_prev. When we call plot()
a custom plot specific to only our class is generated!
plot(prevalence)
Additional Reading
There is a lot more to the world of OOP and many important packages leverage various OOP techniques. In fact, as we mentioned earlier, there are even multiple OOP systems in R. For example, the Tidyverse is built using the S3 system whereas the Bioconductor project uses S4. Of all the available systems, S3 is regarded as the easiest system to learn.
To learn more about OOP, I would highly recommend reading the OOP chapter of
Advanced R and visiting some of the articles
in the {vctrs} package. The
functions used for this post can be found in my {s3examples}
package where I have written a simple S3
subclass of the tibble() class.
-
The term polymorphism has been taken from Advanced R. ↩︎